Context¶
Purpose¶
The goal is to experiment with how to read NorESM data for FAIR2Adapt Case Study 1.
Description¶
In this notebook, we will:
- Open online NorESM data
- Visualize one field
Contributions¶
Notebook¶
- Anne Fouilloux, Simula Research Laboratory (Norway) (reviewer), annefou
Bibliography and other interesting resources¶
Import Libraries¶
# Install xarray-healpy and dggs libraries for regridding
#!pip install git+https://github.com/IAOCEA/xarray-healpy.git git+https://github.com/xarray-contrib/xdggs.git cmcrameri s3fs shapelyimport cartopy.crs as ccrs
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import s3fs # For accessing file storage at Amazon S3
import xarray as xr # N-dimensional arrays with dimension, coordinate and attribute labels
xr.set_options(display_expand_data=False, display_expand_attrs=False, keep_attrs=True)<xarray.core.options.set_options at 0x109e75c10>Open file with Xarray¶
- The file is on the cloud (accessible to everyone)
- The first time, opening the atmosphere ds typically takes a few minutes
- Therefore, we save a local version with selected subset of variables for faster loading the nezxt times
# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = "s3://CS1/data/model/JRAOC20TRNRPv2_hm_sst_2010-01.nc"
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsLoading...
Load grid¶
grid_type = "p"
grid_file = "s3://CS1/data/grid/grid.nc"
grid = xr.open_dataset(s3.open(grid_file))
gridLoading...
Assign coordinates to Xarray Dataset¶
ds = ds.assign_coords({"lon": grid[grid_type + "lon"]})
ds = ds.assign_coords({"lat": grid[grid_type + "lat"]})
dsLoading...
Quick visualization¶
var = "sst"
vmin = -3
vmax = 20
proj = ccrs.NearsidePerspective(
central_longitude=0.0, central_latitude=80.0, satellite_height=3e6
)
fig, ax = plt.subplots(1, figsize=(8, 4.5), dpi=96, subplot_kw={"projection": proj})
p = (
ds[var]
.isel(time=0)
.plot(
ax=ax,
transform=ccrs.PlateCarree(),
x="lon",
y="lat",
add_colorbar=False,
vmin=vmin,
vmax=vmax,
cmap="viridis",
shading="auto",
rasterized=True,
)
)
# Add coastlines and the lat-lon grid
ax.coastlines(resolution="50m", color="black", linewidth=0.5)
ax.stock_img()
gl = ax.gridlines(ylocs=range(15, 76, 15), draw_labels=True)
gl.ylocator = mpl.ticker.FixedLocator([40, 50, 60, 70, 80])
plt.colorbar(p, fraction=0.2, shrink=0.4, label="degC")
ax.set_title("Sea Surface Temperature")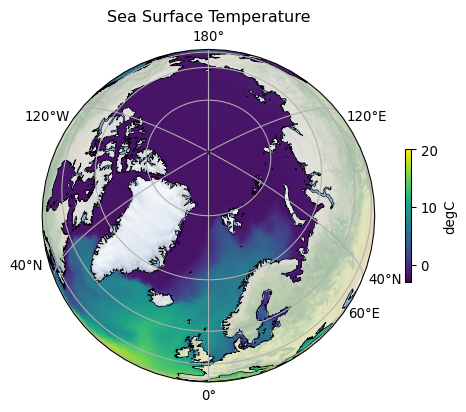
# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = "s3://CS1/data/model/JRAOC20TRNRPv2_hm_U236GF_2010-2018.nc"
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsLoading...
ds.U236GFLoading...
def create_plot(ds, var, vmin, vmax, time=np.nan, sigma=True):
proj = ccrs.NearsidePerspective(
central_longitude=0.0, central_latitude=80.0, satellite_height=3e6
)
fig, ax = plt.subplots(1, figsize=(8, 4.5), dpi=96, subplot_kw={"projection": proj})
if time is not np.nan:
p = (
ds[var]
.isel(time=0)
.plot(
ax=ax,
transform=ccrs.PlateCarree(),
x="lon",
y="lat",
add_colorbar=False,
vmin=vmin,
vmax=vmax,
cmap="viridis",
shading="auto",
rasterized=True,
)
)
elif sigma:
p = ds[var].plot(ax=ax, add_colorbar=False, vmin=vmin, vmax=vmax)
else:
p = ds[var].plot(
ax=ax,
transform=ccrs.PlateCarree(),
x="lon",
y="lat",
add_colorbar=False,
vmin=vmin,
vmax=vmax,
cmap="viridis",
shading="auto",
rasterized=True,
)
# Add coastlines and the lat-lon grid
ax.coastlines(resolution="50m", color="black", linewidth=0.5)
ax.stock_img()
gl = ax.gridlines(ylocs=range(15, 76, 15), draw_labels=True)
gl.ylocator = mpl.ticker.FixedLocator([40, 50, 60, 70, 80])
plt.colorbar(p, fraction=0.2, shrink=0.4, label="degC")
ax.set_title(ds[var].attrs["long_name"])var = "U236GF"
vmin = ds[var].min().values
vmax = ds[var].max().values
print(vmin, vmax)
# create_plot(ds, var, vmin, vmax, sigma=True)# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = "s3://CS1/data/model/JRAOC20TRNRPv2_hm_U236LH_2010-2018.nc"
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsds.U236LH# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = "s3://CS1/data/model/JRAOC20TRNRPv2_hm_U236PPG_2010-2018.nc"
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsds.U236PPG# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = "s3://CS1/data/model/JRAOC20TRNRPv2_hm_U236SF_2010-2018.nc"
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsds.U236SF# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = (
"s3://CS1/data/model/NSSP245frc2NRP205p2v2_hm_I129se40_205001-205912.nc"
)
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsds.I129se40# Define file paths
endpoint_url = "https://server-data.fair2adapt.sigma2.no"
filename_atmosphere = (
"s3://CS1/data/model/NSSP585frc2NRP205p2v2_hm_I129se40_205001-205912.nc"
)
# Extract file from S3
client_kwargs = {"endpoint_url": endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
ds = xr.open_dataset(s3.open(filename_atmosphere))
# Display the subset dataset
dsds.I129se40