Transform NorESM data to DGGS#
Context#
Purpose#
The goal is to experiment with how to transform NorESM data into DGGS (healpix grid).
Warning
This FAIR2Adapt case study will likely require different types of interpolation, as bilinear interpolation is not suitable for all variables. For instance, conservative interpolation is currently being developed.
Description#
In this notebook, we will:
Open online NorESM data
Transform one variable (TREFHT) into DGGS Healpix
Save the transformed data in Zarr
Contributions#
Notebook#
Even Moa Myklebust, Simula Research Laboratory (Norway) (author), @evenmm
Anne Fouilloux, Simula Research Laboratory (Norway) (reviewer), @annefou
Ola Formo Kihle, Independent Consultant / UW Contractor (Norway) (reviewer), @ofk123
Bibliography and other interesting resources#
NorESM dataset used in this notebook, 10.11582/2024.00093
Import Libraries#
# Install xarray-healpy and dggs libraries for regridding
#!pip install git+https://github.com/IAOCEA/xarray-healpy.git git+https://github.com/xarray-contrib/xdggs.git cmcrameri
import os
import numpy as np
import s3fs # For accessing file storage at Amazon S3
import xdggs # Discrete global grid systems in x-array
import xarray as xr # N-dimensional arrays with dimension, coordinate and attribute labels
from xarray_healpy import HealpyGridInfo, HealpyRegridder
xr.set_options(display_expand_data=False, display_expand_attrs=False, keep_attrs=True)
<xarray.core.options.set_options at 0x7813af5349b0>
Open file with Xarray#
The file is on the cloud (accessible to everyone)
The first time, opening the atmosphere ds typically takes a few minutes
Therefore, we save a local version with selected subset of variables for faster loading the nezxt times
# Define file paths
subset_dataset_path = "small_N1850_f19_tn14_20190722.cam.h0.1801-01.zarr"
endpoint_url = 'https://pangeo-eosc-minioapi.vm.fedcloud.eu/'
filename_atmosphere = "s3://afouilloux-fair2adapt/10.11582_2024.00093/N1850_f19_tn14_20190722/atm/hist/N1850_f19_tn14_20190722.cam.h0.1801-01.nc"
# Check if the small dataset exists
if os.path.exists(subset_dataset_path):
print(f"Loading local subset dataset from {subset_dataset_path}")
ds = xr.open_zarr(subset_dataset_path)
else:
# Extract file from S3
print(f"Local subset of variables not found. Loading full dataset from {endpoint_url}")
client_kwargs={'endpoint_url': endpoint_url}
s3 = s3fs.S3FileSystem(anon=True, client_kwargs=client_kwargs)
# Opening the atmosphere ds typically takes a few minutes
atmosphere_ds = xr.open_dataset(s3.open(filename_atmosphere))
# Save a subset for faster access on reload
print("Saving subset of variables to subset_dataset")
atmosphere_ds[["SST", "TREFHT"]].to_zarr(subset_dataset_path)
ds = xr.open_zarr(subset_dataset_path)
# Display the subset dataset
ds
Loading local subset dataset from small_N1850_f19_tn14_20190722.cam.h0.1801-01.zarr
<xarray.Dataset> Size: 113kB
Dimensions: (time: 1, lat: 96, lon: 144)
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* time (time) object 8B 1801-02-01 00:00:00
Data variables:
SST (time, lat, lon) float32 55kB dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
TREFHT (time, lat, lon) float32 55kB dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
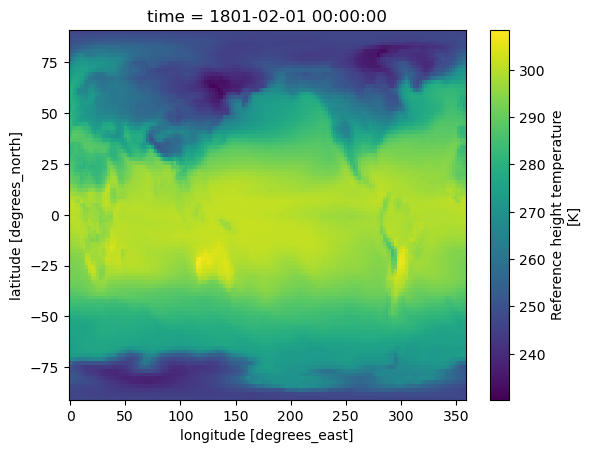
Attributes: (9)Quick visualization of reference height temperature (TREFHT)#
# Plot reference height temperature (TREFHT)
ds.TREFHT.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x78133dbe8260>
Standardize coordinate names and center longitude#
ds = ds.rename_dims({"lat": "latitude", "lon": "longitude"})
ds = ds.rename_vars({"lat": "latitude", "lon": "longitude"})
ds.latitude.attrs["standard_name"] = 'latitude'
ds.longitude.attrs["standard_name"] = 'longitude'
Center around 0 longitude:
ds.coords['longitude'] = (ds.coords['longitude'] + 180) % 360 - 180
ds = ds.sortby(ds.longitude)
Regridding#
Define the target grid (Healpix DGGS)#
def center_longitude(ds, center):
if center == 0:
centered = (ds.longitude + 180) % 360 - 180
else:
centered = (ds.longitude - 180) % 360 + 180
return ds.assign_coords(longitude=centered)
nside = 16 # Each side of the original 12 faces in Healpix is divided into nside parts
healpy_grid_level = int(np.log2(nside)) # Healpix level
number_of_cells = 12*nside**2 # The resulting total number of cells
print("nside:", nside)
print("Level:", healpy_grid_level)
print("Number of cells:", number_of_cells)
nside: 16
Level: 4
Number of cells: 3072
# Define the target Healpix grid
grid = HealpyGridInfo(level=healpy_grid_level)
target_grid = grid.target_grid(ds).pipe(center_longitude, 0).drop_attrs(deep=False).dggs.decode({"grid_name": "healpix", "nside": nside, "indexing_scheme": "nested"})
%%time
# Compute interpolation weights for regridding diff data
regridder = HealpyRegridder(
ds[["longitude", "latitude"]].compute(),
target_grid,
method="bilinear",
interpolation_kwargs={},
)
print(regridder)
HealpyRegridder(method='bilinear', interpolation_kwargs={})
CPU times: user 3.59 s, sys: 228 ms, total: 3.82 s
Wall time: 3.83 s
%%time
regridded = regridder.regrid_ds(ds).pipe(xdggs.decode)
CPU times: user 1.38 s, sys: 616 ms, total: 1.99 s
Wall time: 2.01 s
%%time
ds_regridded = regridded.TREFHT.compute().squeeze()
CPU times: user 186 ms, sys: 12.1 ms, total: 198 ms
Wall time: 196 ms
ds_regridded.dggs.explore()
Save the transformed data to zarr#
url = f"./NorESM-TREFHT-healpix-lvl-{healpy_grid_level}.zarr"
print("url:", url)
ds_regridded.to_zarr(url,mode='w')
url: ./NorESM-TREFHT-healpix-lvl-4.zarr
<xarray.backends.zarr.ZarrStore at 0x78132be8eec0>